What is Y-DNA?
Y-DNA is the DNA of the Y chromosome. Humans have 23 pairs of chromosomes. 22 pairs are autosomal chromosomes and the 23rd pair are the sex chromosomes, which determine a person’s gender. Males have one Y chromosome and one X chromosome (XY), while females have two X chromosomes (XX) and no Y chromosome.
How does Y-DNA hold ancestral information?
These are the unique characteristics of Y-DNA which make it ideal for paternal ancestry analyses:
1. Strict paternal inheritance from father to son
2. Low recombination rate, so it remains “unmixed”
3. Fast mutating STR markers enable the tracing of recent paternal lineages
4. Slow mutating SNP markers determine an individual’s ancient ancestry
Paternal inheritance pattern of Y-DNA
Y-DNA is passed down directly from father to son. This means that all males who descend from the same paternal lineage have exactly the same or very similar Y-DNA profiles.
Trace Your Paternal Line
Free ancestry web apps are available to users who have taken the Y-DNA Test.
Already took the test? Get started for FREE!

Low recombination rate of the Y-DNA
We inherit one copy of each autosomal chromosome from each parent. At each generation, these autosomal chromosome pairs can mix together, and parts of each chromosome can be swapped in a process known as recombination. However, a male only inherits a single copy of the Y chromosome (from their father). This means that there is effectively no chromosome “partner” to mix with, so very little recombination occurs.
Y-DNA has two marker types that provide ancestral information
The Y-DNA has two very different types of markers which are useful for tracing paternal ancestry: STR markers and SNP markers.
1. Y-STR Markers
STR stands for “short tandem repeat.” These are short sequences of DNA (2-13 base pairs) which are repeated over and over again. The number of repeated sequences for each STR marker varies between individuals. This variation is known as an individual’s Y-DNA haplotype or Y-DNA profile.
STR markers have a very fast mutation rate (approximately one mutation every 20 generations). The rapid mutation rate of Y-STR markers makes this marker type useful for examining recent ancestry, i.e. ancestral events within the past few hundred years.
2. Y-SNP Markers
SNP stands for “single nucleotide polymorphism.” This is when a single nucleotide (DNA building block) varies between individuals.
SNP markers have an extremely slow mutation rate (approximately one mutation every few thousand years). SNP markers are used for investigating ancient ancestry from tens of thousands of years ago, and can determine an individual’s Y-DNA haplogroup.
What is the difference between a Y-DNA haplotype and Y-DNA haplogroup?
A Y-DNA haplotype is an individual’s Y-STR profile and includes the number of repeats at specific Y-STR markers. Y-DNA haplotypes are useful for tracing recent paternal lineages and connections.
An individual’s Y-DNA haplogroup represents his “deep ancestry” or ancient family group. Y-DNA studies have shown that all males living today are descendants of a single root paternal ancestor who lived in Africa approximately 150,000 years ago. Over time, our ancestors migrated out of Africa in waves and populated the world. All males can be traced to one of the main Y-DNA haplogroups. Haplogroups are useful for scientists who are studying human migration patterns and have archaeological value. Y-DNA haplogroups are determined by testing Y-SNPs.
Y-STR marker testing to determine Y-DNA haplotype
During Y-STR marker testing, the number of repeats are determined for several different STR markers. The resulting set of repeat numbers is known as the individual’s Y-DNA haplotype (or Y-DNA profile).
The number of repeats for each Y-STR marker does not contain ancestral information by itself. However, it becomes useful when it is compared against other individuals or specific population groups. All males who descend from the same paternal lineage have the same or very similar Y-STR marker profiles. Comparing your Y-STR marker profile against another male in question, or against a database of populations, allows you to search for relatives along your paternal line and also allows you to find out which population groups are the closest match to your paternal lineage.
The next four sections discuss the most common types of Y-STR markers that are used for paternal ancestry testing.
Single-copy Y-STR markers
Single-copy Y-STR markers are STR markers that occur only once in the human genome. A single repeat number (allele value) will show for each single-copy marker in a paternal ancestry report.
Multi-copy Y-STR markers
Multi-copy Y-STR markers are markers that occur more than once in the human genome. More than one allele value will show for each of these markers in a paternal ancestry report.
For example, the DYS385, DYS459 and YCAII are typically present at two different locations on the Y chromosome; therefore, they are also termed “duplicated markers.”
Special multi-copy markers:
- DYS389: Unlike other multi-copy Y-STR markers, only one location of DYS389 is amplified. The forward primer for DYS389 binds at a specific location on the Y chromosome, whereas the reverse primer binds at two different locations. This yields two PCR products: the shorter DYS389I fragment and the longer DYS389II fragment, so two allele values are always reported for this marker.
- DYS464: This is a special Y-STR marker that is known to have 4 to 7 alleles. Previously, the “genotype” was reported. When the genotype was reported, identical repeats were reported multiple times. However, recent policies implemented by the American Association of Blood Banks mandate that all accredited DNA testing laboratories must report the “phenotype” instead of “genotype” for multi-copy markers, especially if the marker has more than 2 alleles. This means that if an individual has duplicate repeat values, they will only be reported once using the “phenotype” reporting procedure.
Using your Y-DNA haplotype to search for, or verify, family linkages
Your Y-DNA haplotype is the same or very similar to that of all males who descend from the same forefather as yourself. This means that your father, grandfather and great-grandfather along your paternal lineage all carry the same Y-DNA haplotype as you.
Once you have tested your Y-STR markers, you can use your haplotype to search the DNA Reunion database for people who are linked to you on your paternal line.
*Only males can take a Y-DNA Test*
Since only males have Y-DNA, only males can take a Y-DNA test to trace their paternal ancestry. Females wishing to trace their paternal ancestry must test the Y-DNA of a male family member such as a brother, father, uncle, or male cousin along their direct paternal lineage.
What is the Atlantic Modal Haplotype?
Some Y-DNA haplotypes occur more frequently in certain parts of the world. For example, people whose ancestors are from the western coast of Europe often share in common a small group of Y-STR markers, which is called the Atlantic Modal Haplotype. The Atlantic Modal Haplotype is tied to the R1b haplogroup and is characterized by these Y-STR markers:
DYS19 = 14
DYS388 = 12
DYS390 = 24
DYS391 = 11
DYS392 = 13
DYS393 = 13
More information about the Atlantic Modal Haplotype can be found in Wilson et al. (2001).
What is genetic distance?
After you test your Y-STR markers, you can compare your markers against the markers of any other male individual to see whether you share a recent common male ancestor. If a match is found, you can determine the TMRCA (time to most recent common ancestor). This is a measurement of approximately how long ago you and the matching individual shared a common ancestor.
A key measurement value when comparing the Y-STR markers between two different individuals is “genetic distance.”
Genetic distance is a measurement of the total difference in allele values of different genetic markers between two individuals. The smaller the value of the genetic distance for a given set of Y-STR markers, the closer two individuals are related, and the more recently they shared a common ancestor. The method used to determine genetic distance for four different Y-STR marker types is explained in the next sections.
Calculation of genetic distance for single-copy and most multi-copy Y-STR markers
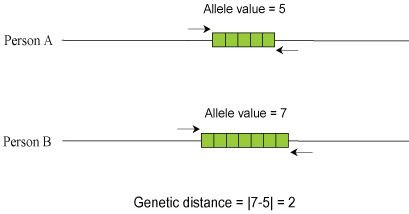
For single-copy STR markers, the calculation is straightforward. The genetic distance for each single copy marker between two individuals is the absolute value of the difference between the value of the markers.
For most multi-copy markers, genetic distance can be calculated by adding the differences in allele values for each of the two copies. The total genetic distance between two individuals is the sum of the genetic distances of all markers compared.
Calculation of genetic distance for multi-copy marker DYS464 – using the “infinite allele” model
Assuming mutations at the same marker took place in a single generation, the “infinite allele” method of determining genetic distance counts the total difference between all copies of the same marker as 1 genetic distance, despite the fact that more than one mutation exists.
The genetic distance for DYS464 is calculated using this method. Regardless of the number of mismatches between two individual at Y-STR marker DYS464, the genetic distance is always reported as a maximum of 1. Please remember that “genetic distance” is not the same as “mismatching markers.”
Calculation of genetic distance for DYS389i/ii
DYS389i is embedded in DYS389ii; therefore, the DYS389i values are included in DYS389ii values. Genetic distance can be determined by adding up two differences: differences in DYS389i values and differences in the second part of DYS389ii values, which are obtained by subtracting the DYS389i values from the DYS389ii values.
What is the TMRCA?
TMRCA stands for “Time to Most Recent Common Ancestor”. It’s a measure of how long ago any two male individuals likely shared a common patrilineal ancestor.
Determining TMRCA through DNA testing:
The TMRCA for any two male individuals can be determined by testing and comparing their Y-STR marker profiles. The more Y-STR markers that are tested and compared, the higher the accuracy of the TMRCA prediction.
Here are some examples of how testing more Y-STR markers can increase the precision of the TMRCA value:
12 Y-STR marker test: If you and someone else test 12 STR markers, and matched each other perfectly at all 12 markers, your TMRCA is approximately 14.4. This means that you and the other individual likely shared a common ancestor between 0 and 14.4 generations ago. Now that’s a very broad time frame and does not provide solid evidence that two individuals are from the same line.
20 Y-STR marker test: If two males match perfectly at 20 STR markers, the TMRCA is narrowed down to 8.3. This means that these two individuals likely descended from the same line and shared a common ancestor anytime between 0 and 8.3 generations ago.
44 Y-STR marker test: If two males match perfectly at 44 STR markers, the TMRCA becomes only 3.8. This means that these two individuals shared a common ancestor between 0 and 3.8 generations ago.
As you can see, the more STR markers that are compared, the more precisely you can narrow down the time frame that you and another person shared a common paternal ancestor.
Why test more Y-STR markers?
A number of different STR markers can be tested in the Y-DNA. The more Y-STR markers that are tested, the more discriminating the matches will be when comparing against other individuals.
For example, a comparison of just 12 Y-STR markers is generally not powerful enough to distinguish family lines and can give inconclusive results. The more markers that are available for comparison, the more discriminating the comparison becomes.
There are two major advantages for comparing more markers:
1. To prevent false positives
2. To obtain conclusive results
Y-STR testing scenario
Mr. Jones has been studying his family’s ancestry for several years and has started a “Jones” family study based in Arizona. He is interested in confirming that his family line is linked to a “Jones” line in New York. Although there are rumours that the two lines are related, Mr. Jones does not have the paperwork to prove this link. Mr. Jones is also interested in finding out whether his line is linked to any other Jones lines worldwide.

Mr. Jones had previously chosen to test just 12 Y-STR markers. After testing, he uses the 12 markers to search the DNA database and finds out that he is a perfect match to the Jones line in New York. However, he also finds that he has a perfect match to over 200 individuals in the DNA Reunion database, and over half of them do not even share his surname. How is this possible? Does it mean that he is related to everyone who matches him at the 12 markers? No, this simply means that data from only 12 markers is not powerful enough to distinguish Mr. Jones from other family lines.
To clarify this, Mr. Jones decides to increase to 20 Y-STR markers. He searches the database using these 20 markers and this time narrows down the number of matches. In fact, now, only 18 people match him perfectly at his 20 markers, including the Jones line in New York. Surprisingly, many of the individuals who matched perfectly at 12 markers, only match at 14 or less out of the 20 markers tested. This confirms that there is no familial link with most of the 200 individuals identified in the 12 marker test, as more than 3 mismatches indicates that two family lines are not closely related.
To further clarify the findings, Mr. Jones decides to upgrade to a 44 marker test. This time, he finds out that he is a perfect match at all 44 markers to only two lines – a Jones line in England, and a Jones line in the United States. After contacting the two lines and comparing paperwork and stories, Mr. Jones was able to confirm that his line was indeed definitely linked to both lines and he is now able to add both new lines to his family tree.
Surprisingly, Mr. Jones was also able to find out that only 43 out of the 44 markers matched with the Jones line in New York. This suggests that although the Jones line in New York is related to his line, they are likely more distantly related.
Mr. Jones also discovered that he had a close match to 4 other Jones lines (43 out of the 44 matched) and he is now pursuing the possibility that the 4 other lines are also distantly related to him. TMRCA analysis dictates that 1 mutation occurs every 500 generations, and thus we would detect a mutation every 12 generations with the 44 marker test.
Mr. Jones is now trying to recruit more Jones males from throughout Europe to try to reconstruct and relink his family line.
Conclusion:
A comparison using only 12 markers was not discriminating enough for Mr. Jones to pinpoint his family lines. After increasing to 20 markers, Mr. Jones was able to obtain more useful information, and was able to eliminate false matches initially generated when only 12 markers were compared. However, after increasing to 44 markers, Mr. Jones was able to pinpoint the people that he was looking for and was furthermore able to accurately answer his questions about his relationship to the Jones line in New York. Mr. Jones can continue to carry on his research, and as more and more people are tested and added to the DNA Reunion database, Mr. Jones will be able to reconstruct his family line in greater detail and re-unite with Jones worldwide who are descendants of his family line.
Tracing ancient ancestry with Y-DNA haplogroups
DNA studies have shown that all people living today can trace their ancestry back to common roots in Africa approximately 150,000 years ago. Over time, man eventually journeyed out of Africa. In many migration waves spanning tens of thousands of years, man eventually populated the rest of the world. During these ancient journeys, SNP mutations occurred randomly in their Y-DNA. Each SNP acts as a “time-and-date stamp” which allows us to understand the approximate time and location in the journey our ancestors were when the SNP first occurred. Once a SNP occurs, it is passed down to all future generations and serves as a marker. This allows us to approximate where our ancestors were at specific time points every few thousand years along the ancient migration out of Africa.
Today, our Y-DNA contains a rich collection of SNP markers, passed down to us from our ancient ancestors over thousands of years. Using SNPs found in our Y-DNA, all people living today can be plotted onto a paternal tree of mankind called the “Y-DNA Phylogenetic Tree.” The main branches of the tree are called “Y-DNA haplogroups.” The finer sub-branches of the tree are called “Y-DNA subclades.”
By testing STR markers in your Y-DNA, you can predict which Y-DNA haplogroup you most likely descended from. However, your Y-DNA haplogroup can only be confirmed by testing SNP markers in your Y-DNA.
Y-DNA haplogroups are associated with different regions of the world
Once you find out which Y-DNA haplogroup you belong to, you can find out which general region of the world your paternal ancestors came from, such as Asia, Europe, Americas (Native American), Africa, or the Middle East.
Haplogroups are NOT country specific because there are no Y-DNA haplogroups that are exclusively found in only one country and not a neighboring country. Y-DNA haplogroups are further classified into Y-DNA subclades, and knowing your subclade may provide further geographical localization of your ancestry.
The following chart shows the Y-DNA haplogroups found in each region.
Region/Population |
Major Y-DNA haplogroups |
| Native Americans | C, Q |
| Oceanic and Aboriginal Australians | C, K, M, N, S |
| East Asian | C, D, N, O, Q |
| South Asian | C, H, L |
| Europe and Middle East | I, J, R, T |
| Diverse | F, G, P |
| African | A, B, E |
A race against time
The scientific community is racing against time to test the DNA of indigenous populations from around the world in order to gain a better understanding of the distribution pattern of Y-DNA haplogroups and subclades found in different parts of the world. This type of study is ongoing and not all Y-DNA subclades currently have distribution data or some may only have limited data.
Y-DNA testing
Y-DNA test options involve testing a panel of STR or SNP markers in the Y chromosome.
Y-DNA Test Type |
Prerequisite |
Purpose |
Description |
| Y-STR Tests | none | For use in searches and comparisons with other individuals | This is the starting point for paternal ancestry research. The options for STR testing include 20, 44 or 101 markers. The 20 marker test is sufficient to use most of the search and analysis features. We recommend initially testing 20 or 44 markers, and then adding more Y-STR markers if you wish to narrow down the matches and TMRCA value. |
| Y-DNA Haplogroup Determination SNP Test | Y-STR Test | To confirm your Y-DNA haplogroup | This test analyzes a selection of SNP markers to confirm which Y-DNA haplogroup you belong to. If you have a “strong” prediction for your Y-DNA haplogroup based on STR testing, your Y-DNA haplogroup may be determined based on STR markers alone. |
| Y-DNA Subclade Determination SNP Test (only available for selected haplogroups) | Y-DNA Haplogroup Determination SNP Test | To confirm your Y-DNA subclade | This test analyzes a selection of Y-SNP markers to confirm your Y-DNA subclade. Subclade testing is currently only available for the following Y-DNA haplogroups: E, G, I, J, L, O, Q, R. |
| Y-DNA Stand Alone SNP Test (only available for selected subclades) | Y-DNA Subclade Determination SNP Test | To further refine your Y-DNA subclade | Over time, new SNPs are discovered and the Y-DNA phylogenetic tree continues to expand. If a new SNP is discovered in your branch of the Y-DNA tree, the new SNP will automatically be offered to you for purchase as a “Stand Alone SNP” test. This allows you to continue to refine your results. |


